Drug Design, Molecular Docking, and ADMET Prediction of CCR5 Inhibitors Based on QSAR Study
TONG Jian-Bo*, ZHANG Xing, BIAN Shuai and LUO Ding
Chin. J. Struct. Chem. 2022, 41, 2202001-2202013 DOI: 10.14102/j.cnki.0254-5861.2011-3268
February 15, 2022
HIV, CCR5 inhibitors, Topomer CoMFA, HQSAR, ADMET
ABSTRACT
The
chemokine receptor CCR5 is a main and necessary co-receptor for which HIV can
recognize and enter the cells, and has been identified as a potential new
target for the design of new anti-HIV therapeutic drugs. Highly active CCR5
inhibitors can prevent HIV-1 from entering target cells and block the process
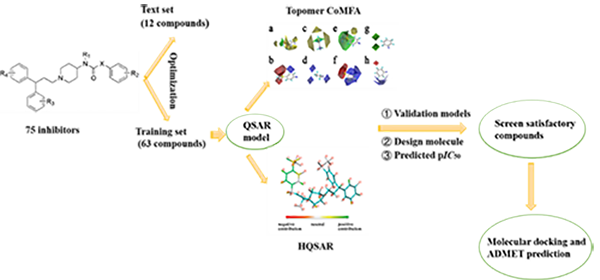
of infection. In this study, HQSAR and Topomer CoMFA methods were used to
establish QSAR models for 75 1-(3,3-diphenylpropyl)-piperidinyl and urea
derivatives, and cross-validation and non-cross-validation were performed on
the generated models. Two models with good statistical parameters and reliable
prediction capabilities are obtained: (Topomer CoMFA: q2 = 0.687, r2 = 0.868, r2pred = 0.623;
HQSAR: q2 = 0.781, r2 = 0.921, r2pred = 0.636). Contour maps and color code maps provide a lot of useful information
for determining structural requirements that affect activity. Topomer search
technology was used for virtual screening and molecular design. Surfex-dock
method and ADMET technology were used to conduct molecular docking, oral
bioavailability and toxicity prediction of the designed drug molecules. Results
showed that A/ASN425, A/GLY198 and A/TRP427 may be the potential active
residues of CCR5 inhibitors evaluated in this study, with 40 newly designed
1-(3,3-diphenylpropyl)-piperidinyl and urea derivatives which have the main
ADMET properties and can be used as a reliable anti-HIV inhibitor. These
results provide a certain theoretical basis for the experimental verification
of new compounds in the future.