Discovery of 4-Thiazol-N-(pyridin-2-yl)pyrimidin- 2-amine as Novel Cyclin-dependent Kinases 4 and 6 Dual Inhibitors via 3D-QSAR and Molecular Simulation
FU Le, ZHAO Li-Nan, GUO Hong-Mei, YU Na, QUAN Wen-Xuan, CHEN Yia, SHU Mao*, WANG Rui and LIN Zhi-Hua*
Chin. J. Struct. Chem. 2022, 41, 2203108-2203124 DOI: 10.14102/j.cnki.0254-5861.2011-3270
March 15, 2022
cyclin-dependent kinases 4 and 6 dual inhibitors, 3D-QSAR, drug design, molecular simulation
ABSTRACT
Cyclin D dependent kinases 4/6 regulate the entry of cells into S phase
and are effective target for the discovery of anticancer drugs. In this
article, 3D-QSAR modeling including
comparative molecular field analysis (CoMFA) and comparative molecular
similarity indices analysis fields (CoMSIA) was implemented on 52 dual CDK4/6
inhibitors. As a result, we obtained a pretty good 3D-QSAR model, which is CoMFACDK4 with q2 to be 0.543 and r2 to be 0.967; CoMSIACDK4 with q2 being 0.518
and r2 being 0.937; CoMFACDK6 with q2 to be 0.624 and r2 to be 0.984; CoMSIACDK6 with q2 being
0.584 and r2 being 0.975.
Molecular docking confirmed the important residues for interactions. Molecular
dynamics simulation further confirmed binding affinity with key residues of
protein, such as Lys22, Lys35, Val96 for CDK4 and Lys43, His100, Val101 for
CDK6 at the active sites. Then these results offered new directions to explore
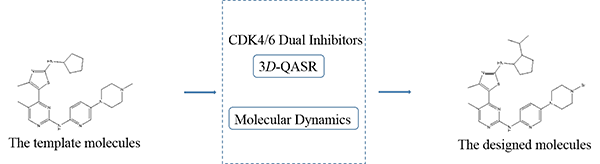
new inhibitors of CDK4/6. Finally, we designed 10 novel compounds with
promising expected activity and ADME/T properties, and provided referable
synthetic routes.